CONP Portal | Dataset
Hippocampal morphology and cytoarchitecture in the 3D BigBrain
| Other Dates: | Start Date: 2019-04-01 00:00:00 |
|---|
Description:
Dataset README information
BIDS_40um/sourcedata/ contains the original left and right hippocampal block nifti images (ftp://bigbrain.loris.ca/BigBrainRelease.2015/2D_Final_Sections/Coronal/Png/Full_Resolution/), which is publically available under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International Public License (https://bigbrain.loris.ca/main.php?test_name=license). BIDS_downsampled/sourcedata contains 80um resampled blocks, on which manual labelling of hippocampal and surrounding structures was performed (BIDS_downsampled/manual_masks/).
Laplacian solutions were obtained over the domain of hippocampal grey matter according to our previous publication (https://www.ncbi.nlm.nih.gov/pubmed/29175494), code available at https://github.com/jordandekraker/HippUnfolding (BIDS_downsampled/HippUnfold). These solutions were obtained in the downsampled image space due to memory limits, and were then upsampled via the upsample.m script, which populated the corresponding directories in BIDS_40um.
In parallel, an equivolume laminar solution was also obtained over the domain of hippocampal grey matter using code available at https://github.com/nighres/nighres. The exact code used to obtain these solutions can be found in BIDSscripts/nighres.
All analyses performed in the current publication can be replicated using the scripts found in BIDSscripts. Many of these scripts depend on the outputs from other scripts. For example, allfeatures.m collects, preprocesses, and plots the 5 morphological features computed in MorphologyFeatures.m as well as the 10 laminar features computed in LaminarFeatures.m. For ease of use, .mat files are provided alongside each script containing all variables needed to run downstream analyses.
BIDSscripts/figures contains visualizations of all analyses, as well as supplementary analyses and some which were not included with our publication because no conclusive assertions could be made form them.
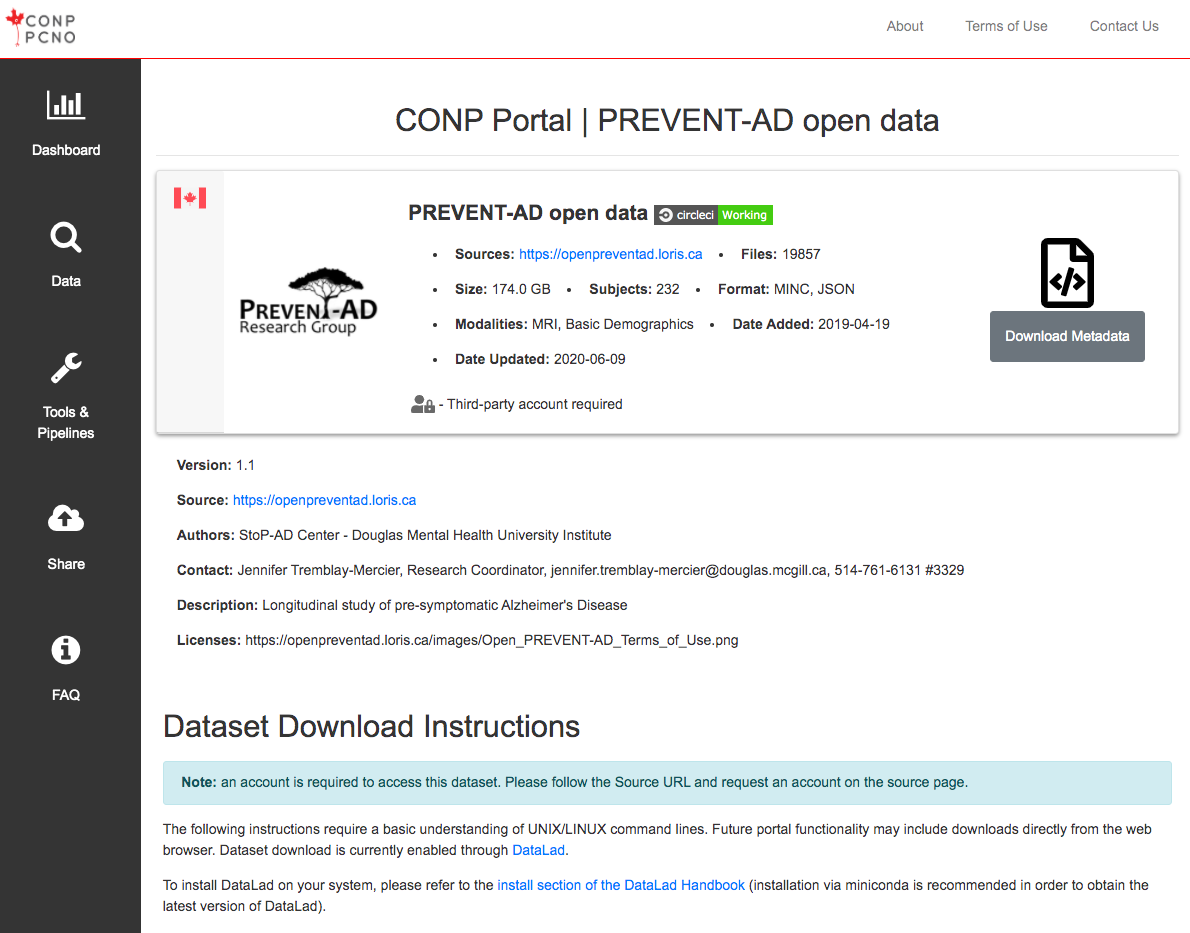
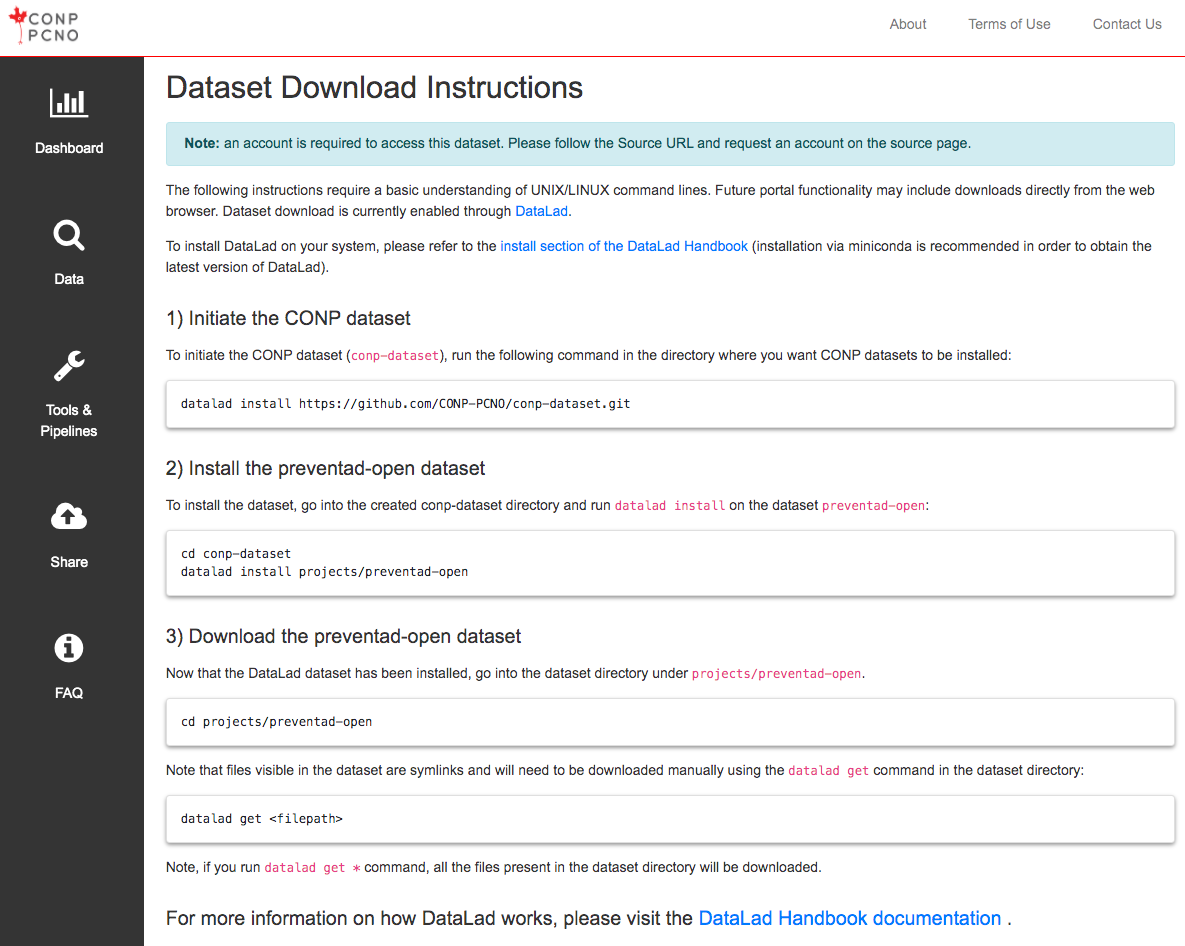
The following instructions require a basic understanding of UNIX/LINUX command lines. Future portal functionality may include downloads directly from the web browser. Dataset download is currently enabled through DataLad.
Note: The conp-dataset requires version >=0.12.5 of DataLad
and version >=8.20200309 of git-annex.
To install DataLad on your system, please refer to the install section of the DataLad Handbook (installation via miniconda is recommended in order to obtain the latest version of DataLad).
1) Initiate the CONP dataset
To initiate the CONP dataset (conp-dataset), run the following
command in the directory where you want CONP datasets to be installed:
datalad install https://github.com/CONP-PCNO/conp-dataset.git
2) Install the Khanlab/BigBrainHippoUnfold dataset
To install the dataset, go into the created conp-dataset directory and run
datalad install on the dataset Khanlab/BigBrainHippoUnfold:
cd conp-dataset
datalad install projects/Khanlab/BigBrainHippoUnfold
3) Download the Khanlab/BigBrainHippoUnfold dataset
Now that the DataLad dataset has been installed, go into the dataset
directory under projects/Khanlab/BigBrainHippoUnfold.
cd projects/Khanlab/BigBrainHippoUnfold
Note that files visible in the dataset are symlinks and will need to be
downloaded manually using the
datalad get
command in the dataset directory:
datalad get <filepath>
Note, if you run datalad get * command, all the files present
in the dataset directory will be downloaded.