CONP Portal | FAQ
CONP | Data Access & Download | Data Upload | Tools & Pipelines | DataLad
I. The Canadian Open Neuroscience Platform (CONP)
What is the CONP?
The Canadian Open Neuroscience Platform (CONP) is a national network comprising organizational, scientific, and technical infrastructure to support:
- open, global sharing of multi-modal neuroscience data and advanced analytic tools
- scholar training in the cross-disciplinary use of Open Neuroscience practices
- creation of policy frameworks for ethically sound data governance and open dissemination
- the growth of Open Publishing and its facilitation of reproducibility by the wider scientific community
- building partnerships with international initiatives with similar Open Science principles and objectives.
What is the CONP Portal?
The CONP Portal aims to facilitate the FAIR sharing of data and analysis tools through standard and open technologies. Features include:
- Multiple data modalities (fMRI, PET, genomic, etc.)
- Multiple access tiers
- Flexible, federated data model
- Common metadata crucial to FAIR principles and provenance tracking
- Browser-based download for completely open data
- Data access via DataLad for either open or registered datasets
- Access to high-performance computing through CBRAIN
II. Data Access and Download
What data are contained in the CONP Portal?
The CONP Portal contains a diverse range of datasets, primarily neuroimaging, but also transcriptomics, genomics, and other related data modalities. Many of these datasets come from neuroscience research institutes, while others link to public resources that may be of interest to neuroscientists. A full list can be found here.
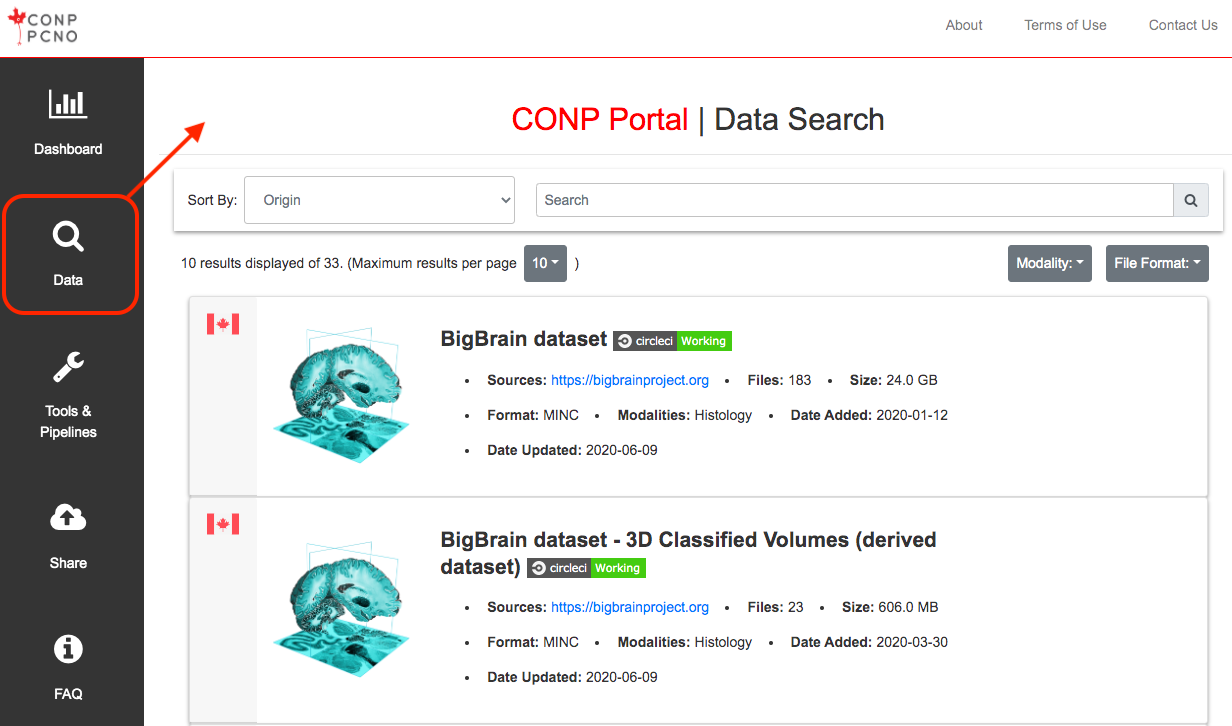
How do I download data from the CONP Portal?
Individual dataset pages accessible from the search page contain instructions for downloading data. At the moment, access to datasets is made available through either a one-click, browser-based download function, or through the DataLad data management system, which should be version 0.12.2 or higher (installation details). Please refer to this tutorial for a step-by-step guide to downloading datasets from the CONP Portal.
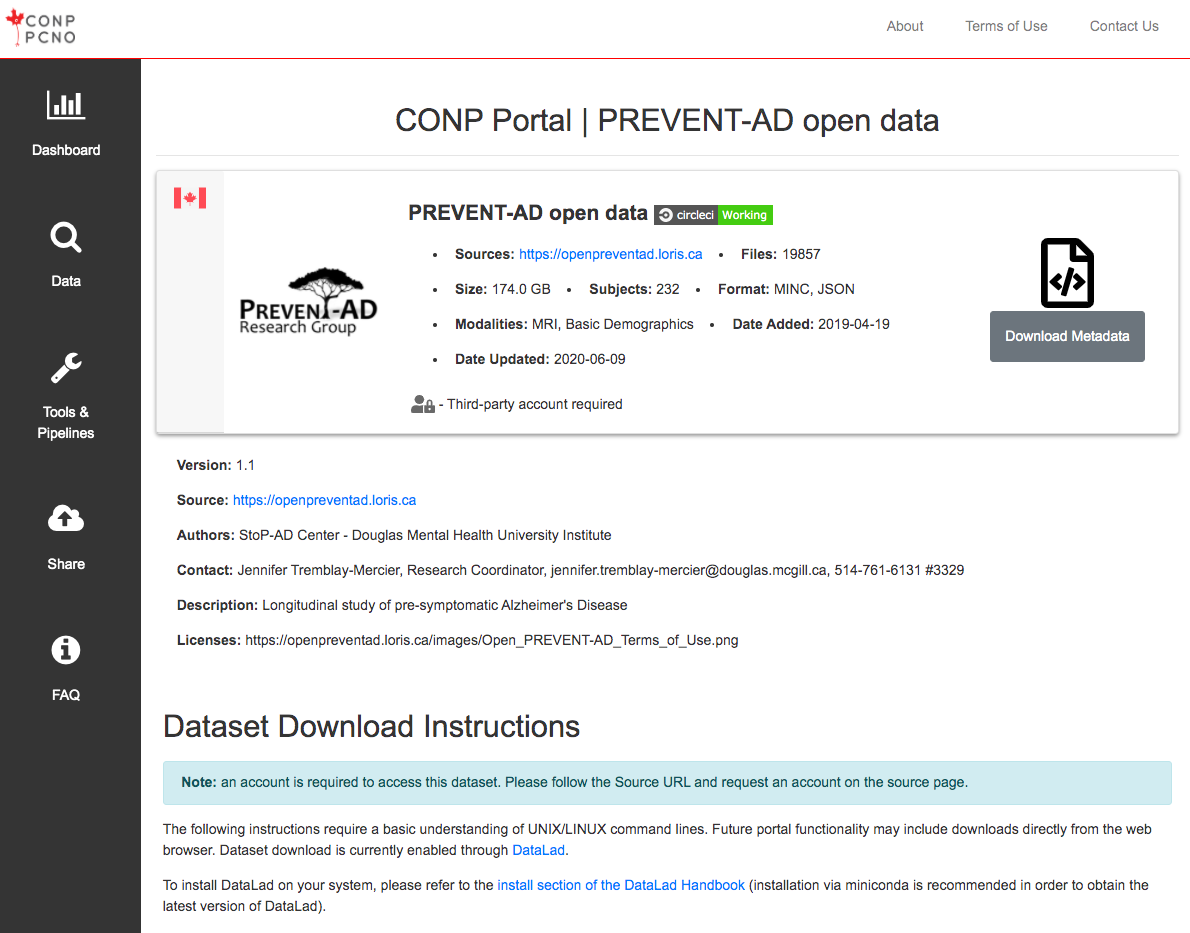
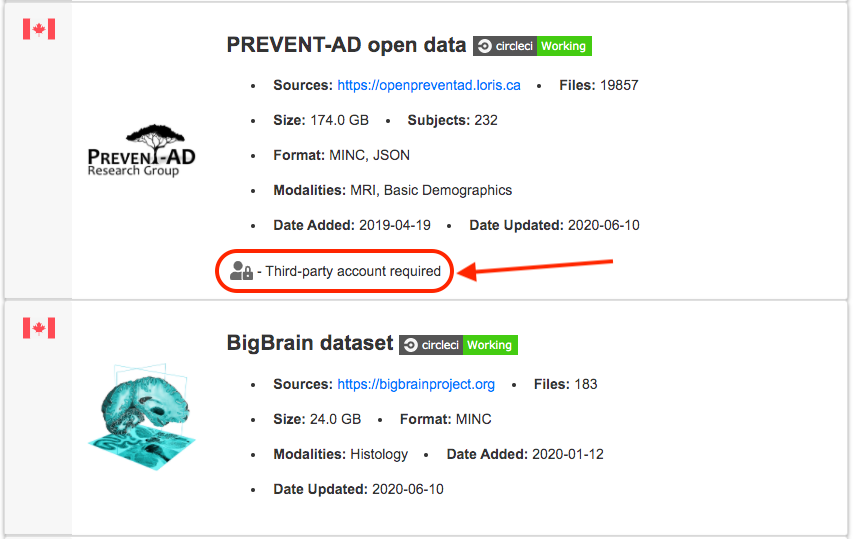
How do I access restricted/private datasets?
If a dataset requires a third-party account, please follow the link embedded in the “Third-party account required” text at the top-right of the dataset page for further information.
III. Data Sharing/Upload
Why should I upload data to the CONP Portal?
The CONP Portal brings together datasets and analysis tools from different sources by harmonizing their metadata and, in many cases, offering direct access to data, as opposed to merely indexing its availability from another site. This allows users to search across these different sources to find related data and tools that are normally stored separately.
This is key to facilitating discoverability, reuse, and deeper scientific exploration of data. For example, it allows a researcher to discover multiple sources of data responding to given search criteria (e.g., according to modality, participant population, etc.), retrieve those data either through the Portal’s web interface or through command-line access, and apply containerized tools to these data either locally or through Cloud computing. These features increase your data’s visibility, accessibility, and utility to the neuroscience community.
How do I upload data to the CONP Portal?
Data can be uploaded to the CONP Portal either through Zenodo by tagging your dataset with the keyword canadian-open-neuroscience-platform, through OSF by tagging your dataset with canadian-open-neuroscience-platform and setting the dataset to Public, to the CONP Community Server, or manually through DataLad. Please see detailed instructions for these options by clicking the “Share” button at the left of your screen.
IV. Tools & Pipelines
What tools/pipelines are contained in the CONP Portal?
The Portal contains a diverse range of analytical tools and pipelines. Many of these come from neuroscience or genomics research institutes. A full list can be found here.
What are the requirements to install and run a tool?
CONP pipelines can be easily installed and run on any computer with a container engine. You will need to install:
- Docker or Singularity
- Boutiques python package (
pip install boutiques)
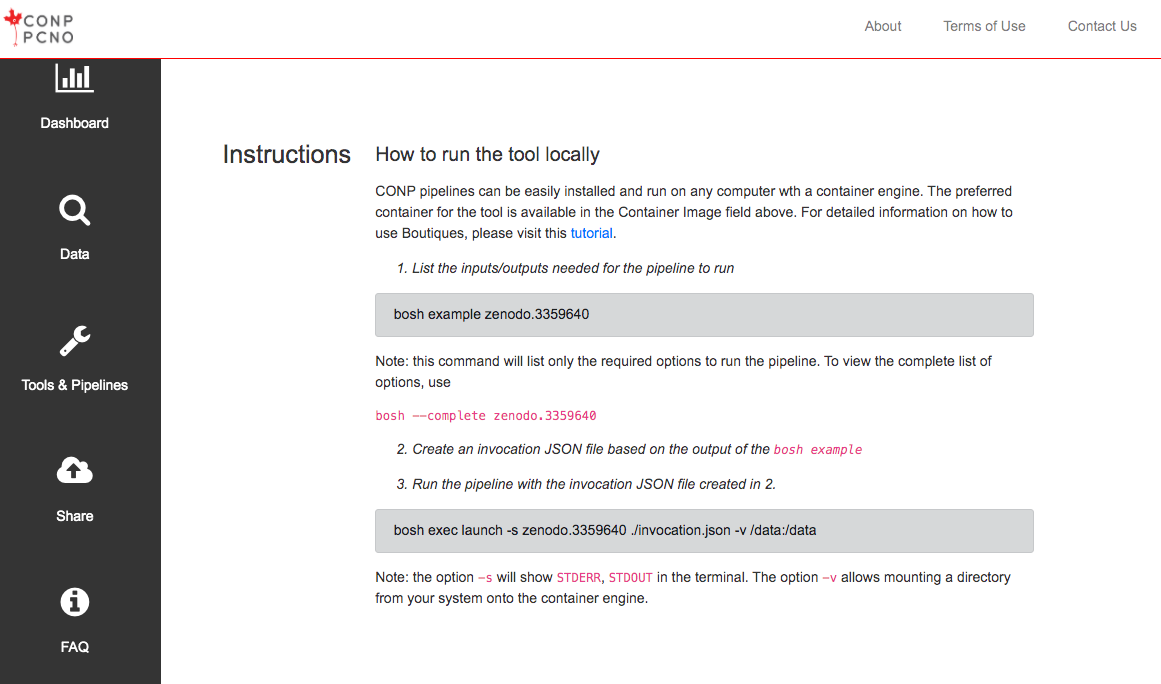
How can I run a CONP tool/pipeline?
To run tools/pipelines locally, make sure your have Boutiques installed alongside either Docker or Singularity.
If you have never used Boutiques before, we recommend this tutorial.
Tools/pipelines can be run on Boutiques based on their Zenodo ID as follows:
-
The example command will create a first minimal invocation so that you don't have to start from scratch:
bosh example <zenodo_id>. If you feel like starting with a more complete set of options, you can pass--completeto the example command:bosh example --complete <zenodo_id> -
You can now edit a JSON example invocation (
example_invocation.json) to add your input values based on the example provided in 1) -
You are now all set to use the
execcommand to launch an analysis. Note, the option-swill show livestdout/stderrin the terminal.
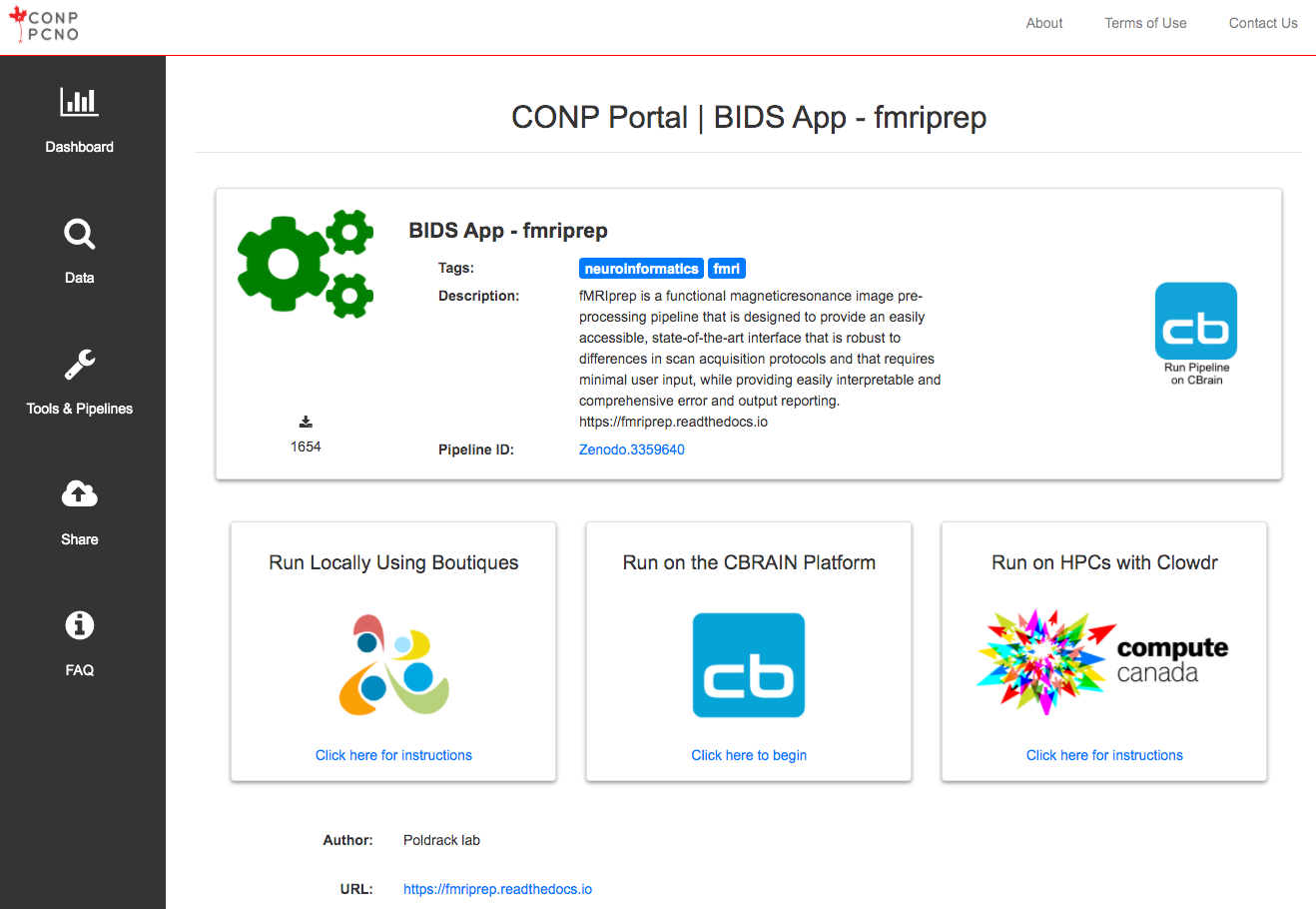
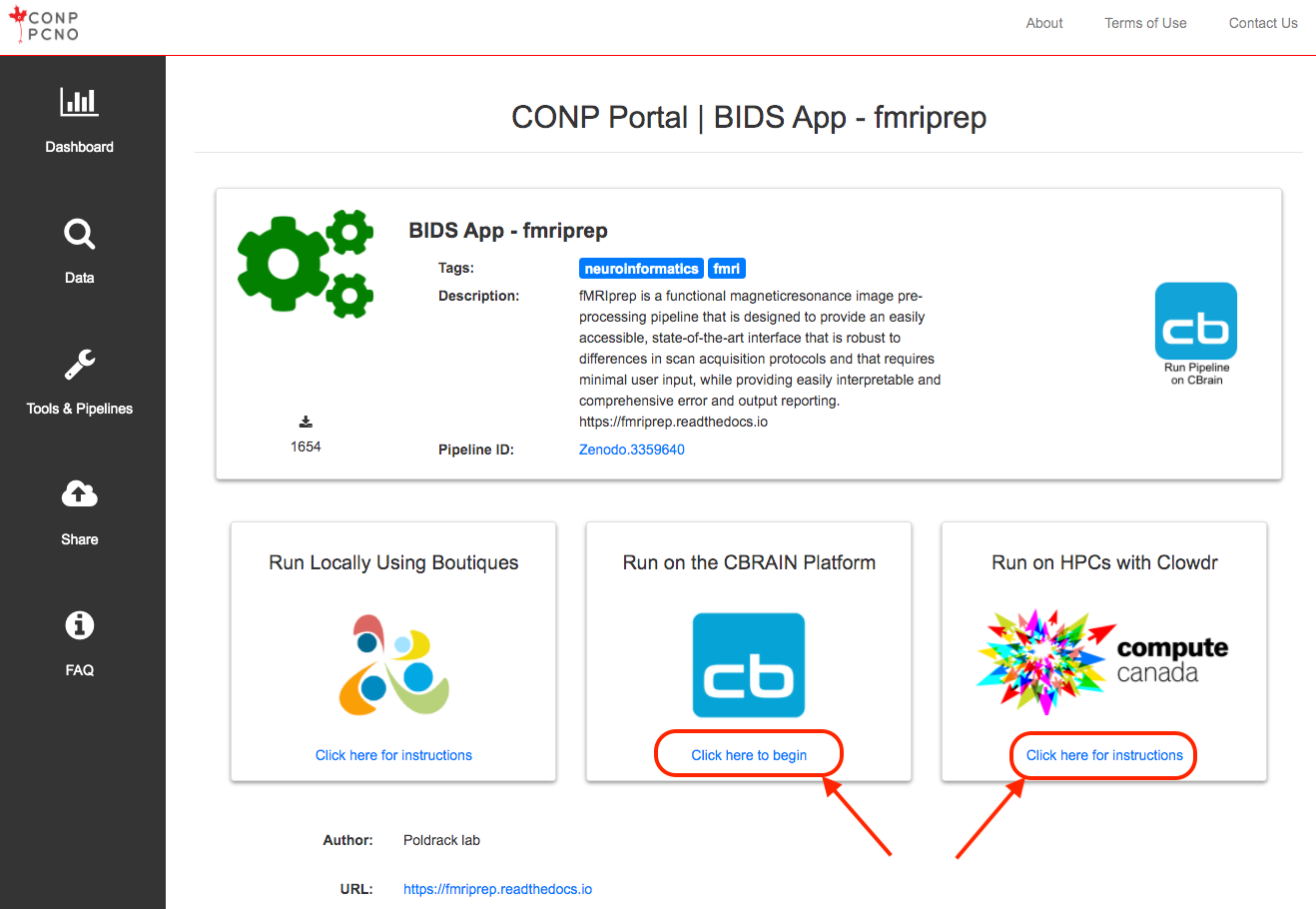
bosh exec launch -s <zenodo_id> <invocation>Note that in addition to the ability to run the tools/pipelines locally, it is possible to run some tools/pipelines via the CBRAIN platform (for example: the fmriprep BIDS App, Zenodo.3359640). Clicking on the blue icon "Use This Tool On CBRAIN" will bring you to the CBRAIN page where you can run pipelines on the super-computing network administered by the Digital Research Alliance of Canada. For more information on CBRAIN, visit http://www.cbrain.ca/.
I get the following error when running the tool on bosh
Cannot connect to the Docker daemon at unix:///var/run/docker.sock. Is the docker daemon running?
docker: Cannot connect to the Docker daemon at unix:///var/run/docker.sock. Is the docker daemon running?.
See 'docker run --help'.
...
Exit code
125=> ensure you have Docker installed and running before executing the bosh command.
</data/dataset> does not exist
Traceback (most recent call last):
File "/run.py", line 53, in <module>
run('bids-validator %s'%args.bids_dir)
File "/run.py", line 25, in run
raise Exception("Non zero return code: %d"%process.returncode)
Exception: Non zero return code: 2
...
Exit code
1=> confirm your data directory is mounted on the container by using the -v option in the bosh command. For example, if your data is in /data, then you need to specify -v /data:/data in the bosh command as follows:bosh exec launch -s <zenodo_id> <invocation> -v <directory_to_mount>
How can I upload/share my own tool/pipeline on the CONP Portal?
You can add your own tool/pipeline by following the instructions described in this document.
V. DataLad
Installing required software - Method 1: NeuroDebian
The following instructions assume a relatively recent Ubuntu- or Debian-based Linux system, though equivalent steps can be taken with other operating systems.
One of the most convenient ways of installing a host of neuro-related software is through the Neurodebian repository. For the following installation procedure, we will assume the use of NeuroDebian, which is installed with:
sudo apt-get update
sudo apt-get install neurodebian
Python
The CONP recommends you use Python version 3.12 and up. You can check your Python version with:
python --versionor python3 --version
git
sudo apt-get install git
It is strongly recommended that you configure your git credentials to avoid having to enter them repeatedly or other complications when using DataLad:
git config --global user.name "Jane Doe"
Replace "Jane Doe” with your name.
git config --global user.email “janedoe@example.com”
Replace “janedoe@example.com” with your email address.
git-annex
sudo apt-get install git-annex-standalone
As of late 2024, this installs git-annex version 10.20241202 or above, which works with CONP datasets. Earlier versions are also compatible but the CONP recommends a relatively recent version for maximum ease. The version of git-annex installed can be verified with:
git-annex version
DataLad
sudo apt-get install datalad
Installing required software - Method 2: pip
The same requirements for Python and git apply from the previous section.
Recent versions of Ubuntu-based distributions (e.g., based on 24.04 LTS) require virtual environments for external Python packages. You can use venv or pipx to do this. The easiest for most users is probably to use pipx, in which case git-annex and datalad are installed with the following commands:
pipx install datalad-installer
datalad-installer git-annex -m datalad/git-annex:release
git config --global filter.annex.process "git-annex filter-process"
pipx install datalad
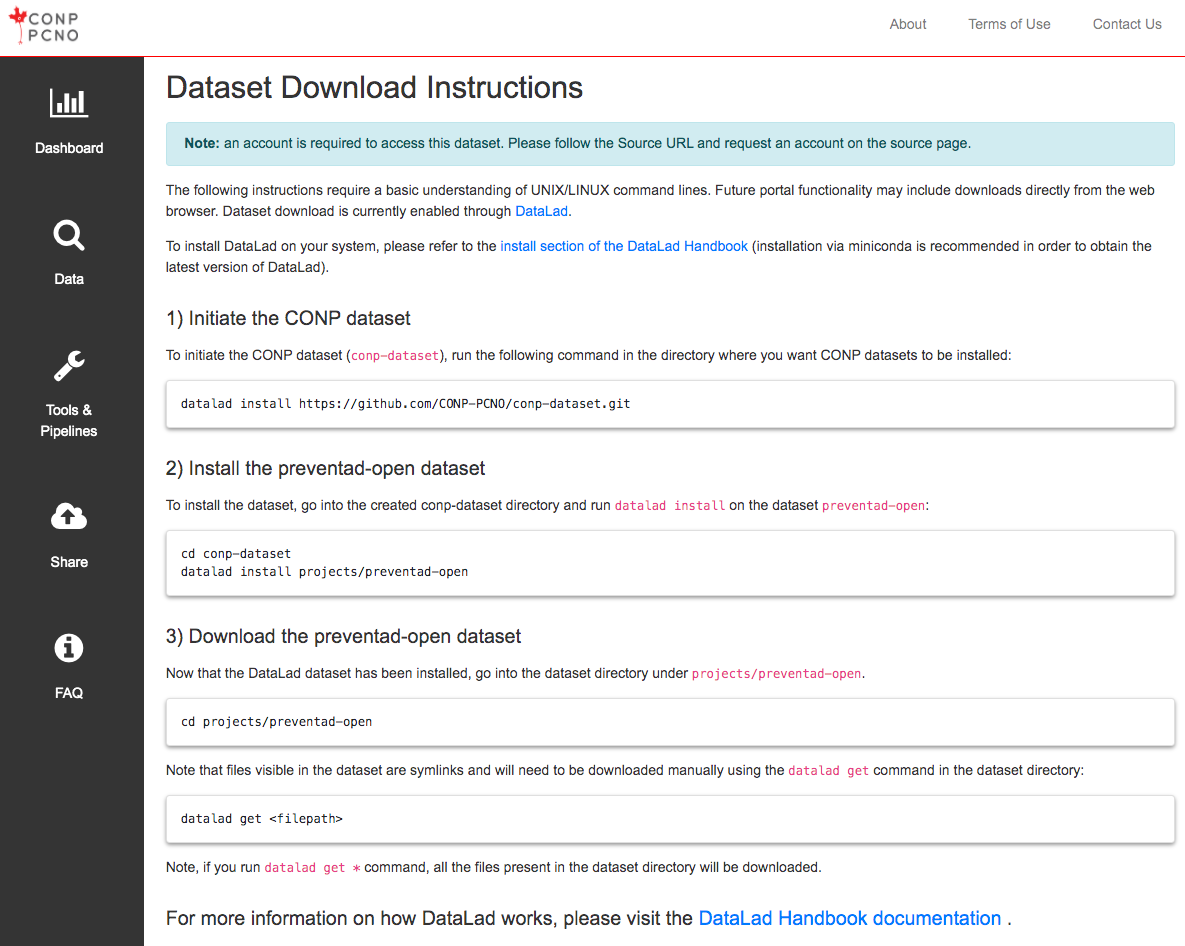
To which username/password is DataLad referring to when trying to download via datalad get preventad-open or multicenter-phantom datasets?
In order to have access to those datasets, you will need to request an account on their respective LORIS repositories.
To do so, on the dataset page, click on the link shown under "Source" which will redirect you to the LORIS login page. Below the login button there is a 'Request Account' link that will take you to a Request Account form that will need to be filled and submitted.
Once the account is approved, you will receive an email with the login credentials that you can use to either browse the LORIS repository or download data via DataLad using the datalad get command.
How can I update a dataset to benefit from the latest version?
From the conp-dataset root directory, run the following command to update all the datasets:
datalad update --merge 
 Experiments - Beta
Experiments - Beta