CONP Portal | Dataset
Accurate registration of the BigBrain dataset with the MNI PD25 and ICBM152 atlases
| Other Dates: | Start Date: 2019-03-19 00:00:00 |
|---|
Description:
Dataset README information
This dataset includes co-registration of the BigBrain dataset to the MNI PD25 atlas and the ICBM152 2009b atlases.
The data include deformed BigBrain atlases and manual subcortical segmentations in MINC2 and NIFTI-1 formats, as well as relevant spatial transformation in MINC transformation format.
The segmented subcortical structures include: red nucleus, subthalamic nucleus, substantia nigra, caudate, putamen, globus pallidus externa, globus pallidus interna, thalamus, hippocampus
Note that the described improved co-registration was performed upon the BigBrain data in ICBM space from the BigBrain 2015release.
Within this dataset, the down-sampled versions of BigBrain atlases are distributed under the CC BY4.0 License upon the consent from the original data owners, the Montreal Neurological Institute
(Montreal, Canada) and the Forschungszentrum Jülich (Jülich, Germany). However, this exception to the existing BigBrain dataset does not alter the general term of that license for use of the
BigBrain itself, which is still under the CC BY-NC-SA 4.0 License.
The included files are as follows:
-
Deformed BigBrain atlases:
BigBrain in PD25 space: BigBrain-to-PD25-nonlin-{300um, 0.5mm, 1mm}
BigBrain in ICBM152 symmetric atlas: BigBrain-to-ICBM2009sym-nonlin-{300um, 0.5mm, 1mm}
BigBrain in ICBM152 asymmetric atlas: BigBrain-to-ICBM2009asym-nonlin-{300um, 0.5mm, 1mm}
Synthetic T2w PD25 atlas: PD25-SynT2-template-{300um, 0.5mm, 1mm}
T1-T2* fusion PD25 atlas: PD25-enhanceFusion-template-{300um, 0.5mm, 1mm} -
Manual subcortical segmentations:
BigBrain coregistered to ICBM in the BigBrain2015 release: BigBrain-segmentation-0.3mm
MNI PD25: PD25-segmentation-0.5mm
ICBM152 2009b symmetric: ICBM2009b_sym-segmentation-0.5mm
ICMB152 2009b asymmetric: ICBM2009b_asym-segmentation-0.5mm -
Related transformations:
BiBrain-to-PD25: BigBrain-to-PD25-nonlin.xfm
BigBrain-to-ICBM2009asym: BigBrain-to-ICBM2009asym-nonlin.xfm
BigBrain-to-ICBM2009sym: BigBrain-to-ICBM2009sym-nonlin.xfm
PD25-to-ICBM2009asym: PD25-to-ICBM2009asym-nonlin.xfm
PD25-to-ICBM2009sym: PD25-to-ICBM2009sym-nonlin.xfm -
List of subcortical labels: subcortical-labels.csv
References:
For the methods used, please cite the following articles:
-
Y. Xiao, J.C. Lau, T. Anderson,J. DeKraker, D. Louis Collins, T.M. Peters, and A.R. Khan, “Bridging micro and macro:
accurate registration of the BigBrain dataset with the MNI PD25 and ICBM152 atlases,” bioRxiv, 561118, Cold Spring Harbor Laboratory,
doi: https://doi.org/10.1101/561118. -
Y. Xiao, V. Fonov, S. Beriault, F.A. Subaie, M.M. Chakravarty, A.F. Sadikot,
G. Bruce Pike, and D. Louis Collins, “Multi-contrast unbiased MRI atlas of a
Parkinson's disease population,” International Journal of Computer-Assisted
Radiology and Surgery, vol. 10(3), pp. 329-341, 2015.
3.Y. Xiao, V. Fonov, S. Beriault, F.A. Subaie, M.M. Chakravarty, A.F. Sadikot, G. Bruce Pike, and D. Louis Collins,
“A dataset of multi-contrast population-averaged brain MRI atlases of a Parkinson’s disease cohort,”Data in Brief, 2017.
When using the downsamled BigBrain atlases, please cite the following article:
- Amunts, K. et al.: “BigBrain: An Ultrahigh-Resolution 3D Human Brain Model”, Science (2013) 340 no. 6139 1472-1475, June 2013
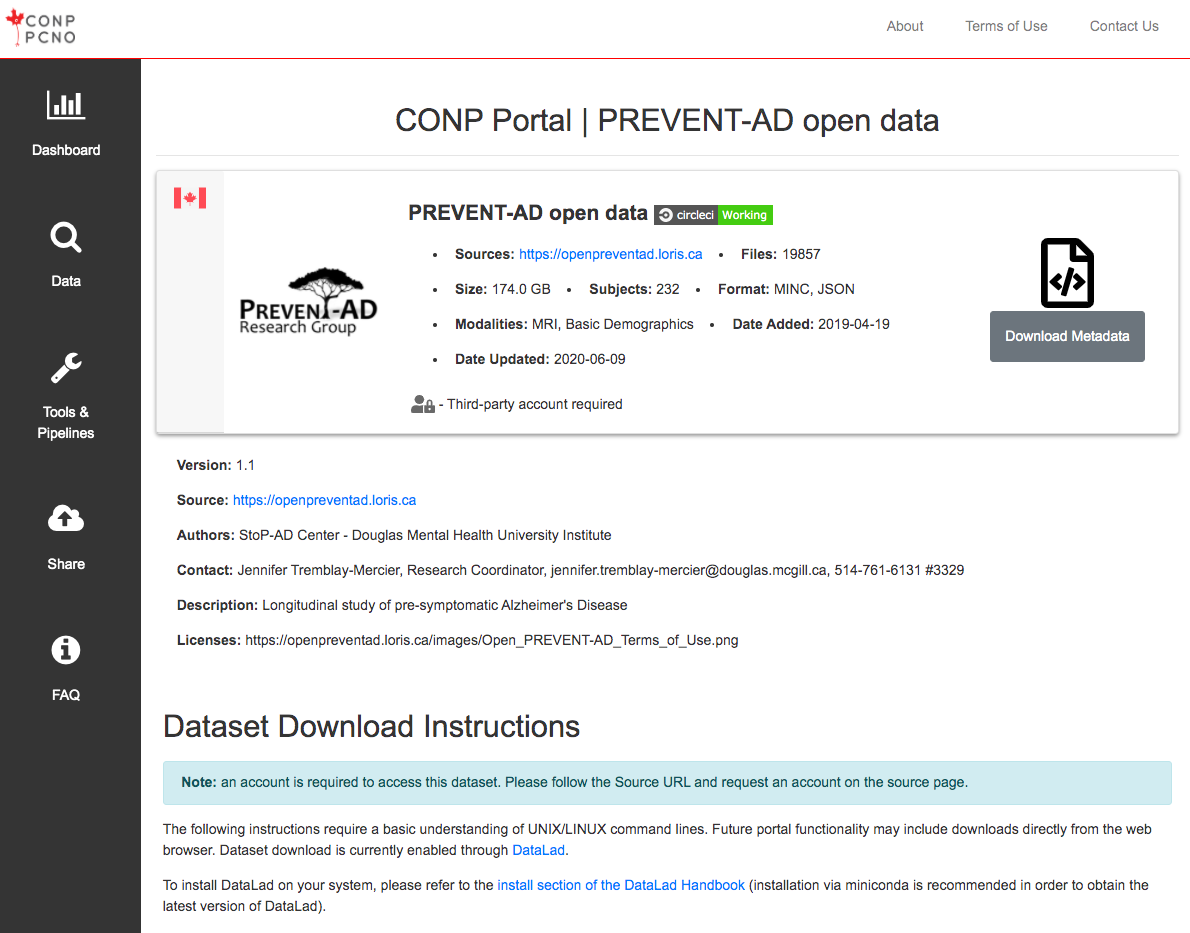
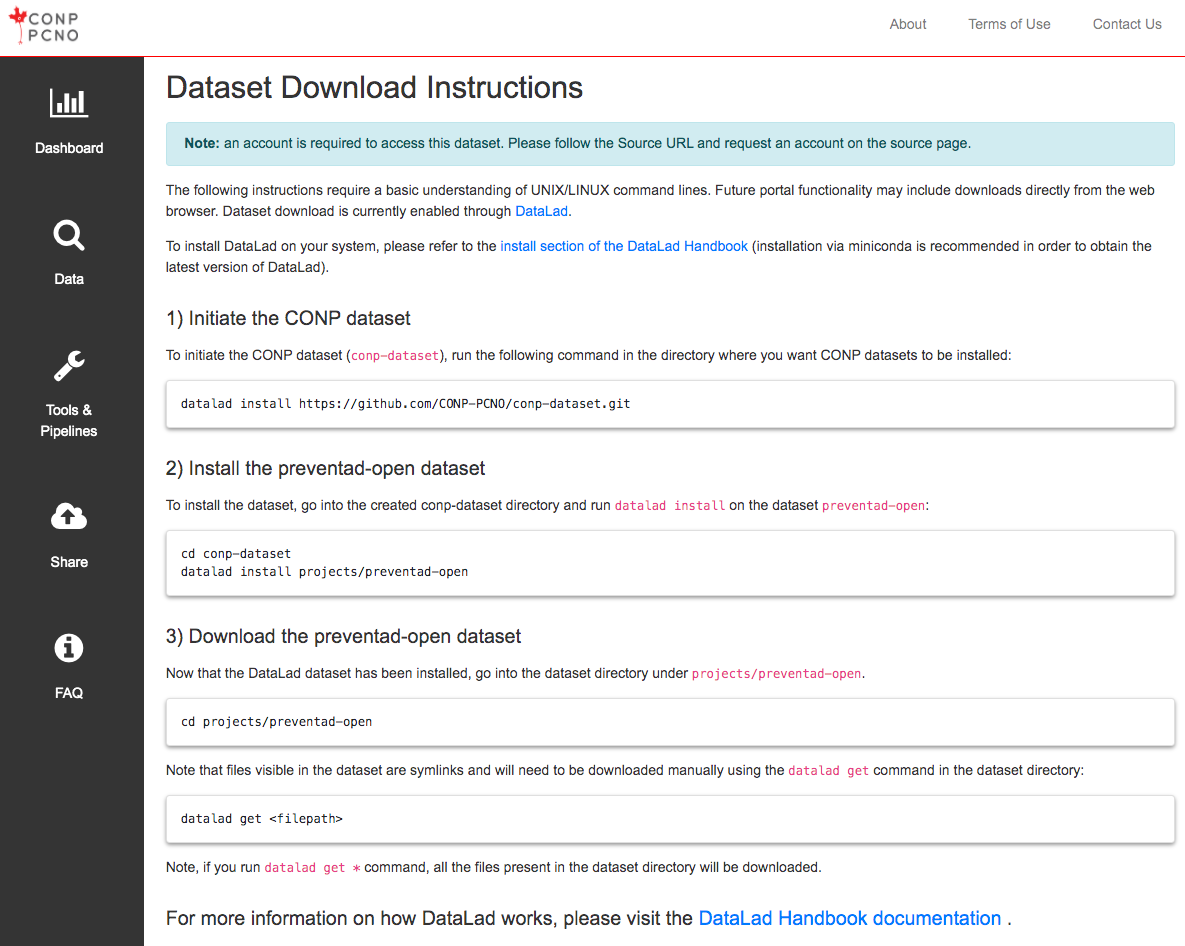
The following instructions require a basic understanding of UNIX/LINUX command lines. Future portal functionality may include downloads directly from the web browser. Dataset download is currently enabled through DataLad.
Note: The conp-dataset requires version >=0.12.5 of DataLad
and version >=8.20200309 of git-annex.
To install DataLad on your system, please refer to the install section of the DataLad Handbook (installation via miniconda is recommended in order to obtain the latest version of DataLad).
1) Initiate the CONP dataset
To initiate the CONP dataset (conp-dataset), run the following
command in the directory where you want CONP datasets to be installed:
datalad install https://github.com/CONP-PCNO/conp-dataset.git
2) Install the Khanlab/BigBrainMRICoreg dataset
To install the dataset, go into the created conp-dataset directory and run
datalad install on the dataset Khanlab/BigBrainMRICoreg:
cd conp-dataset
datalad install projects/Khanlab/BigBrainMRICoreg
3) Download the Khanlab/BigBrainMRICoreg dataset
Now that the DataLad dataset has been installed, go into the dataset
directory under projects/Khanlab/BigBrainMRICoreg.
cd projects/Khanlab/BigBrainMRICoreg
Note that files visible in the dataset are symlinks and will need to be
downloaded manually using the
datalad get
command in the dataset directory:
datalad get <filepath>
Note, if you run datalad get * command, all the files present
in the dataset directory will be downloaded.