CONP Portal | Dataset
BigBrain dataset - Surface Parcellations (derived dataset)
| Is About: | Homo sapiens, adult |
|---|
Description:
Dataset README information
BigBrain Surface Parcellations
BigBrain
The BigBrain is the brain of a 65 years old man with no neurological or psychiatric
diseases in clinical records at time of death. The brain was embedded in parafin and
sectioned in 7404 coronal histological sections (20 microns), stained for cell bodies.
The BigBrain is the digitized reconstruction of the hi-res histological sections
(20 microns isotropic).
Dataset content
This dataset contains surface parcellations from the BigBrain release 2015
published in the BigBrain Project website.
Commonly used surface atlases previously defined on fsaverage (Brainnetome, DKT, Economo, Schaefer2018) and HCP's fs_LR (MMP 1.0) have been mapped onto the BigBrain. Surface-based registration has been achieved using a customized MSM pipeline.
The data is organized as follows:
- fsaverage folder: contains atlases on fsaverage projected on the resampled BigBrain surfaces in fsaverage space
- fs_LR folder: contains atlases on fs_LR projected on the resampled BigBrain surgaces in fs_LR space.
- BigBrain_space folder: contains resampled labels of the various atlases defined on the original BigBrain surfaces.
Reference and more information
If you are using this dataset, please cite:
Lewis, L.B., Lepage, C.Y., Glasser, M.F., Coalson, T.S., Van Essen, D.C.,
and Evans, A.C. (2019). 'An extended MSM surface registration pipeline to
bridge atlases across the MNI and the FS/HCP worlds', OHBM poster, Rome.
Lewis, L.B., Lepage, C.Y., Glasser, M.F., Coalson, T.S., Van Essen, D.C.,
and Evans, A.C. (2020, submitted). 'An improved MSM surface registration
pipeline to bridge atlases across the MNI and the FS/HCP worlds', OHBM poster,
Montreal / virtual.
The references for the atlases are:
- Brainnetome:
Fan, L. et al. (2016). 'The Human Brainnetome Atlas: a new brain atlas
based on connectional architecture', Cerebral Cortex 26:3508-26.
- DKT:
Desikan, R.S. et al. (2006). 'An automated labeling system for subdividing
the human cerebral cortex on MRI scans into gyral based regions of interest',
NeuroImage 31(3):968-80.
Klein, A., and Tourville, J. (2012). '101 labeled brain images and a consistent
human cortical labeling protocol', Frontiers in Neuroscience 6(171).
- Economo:
Scholtens, L.H. et al. (2018). 'An MRI Von Economo - Koskinas atlas', NeuroImage,
170:249-256.
- Schaefer2018:
Schaefer, A. et al. (2018). 'Local-global parcellation of the human cerebral
cortex from intrinsic functional connectivity MRI', Cerebral Cortex 29:3095-3114.
- HCP:
Glasser, M.F. et al. (2016). 'A multi-modal parcellation of human cerebral
cortex', Nature 536:171-178.
- BigBrain reference:
The BigBrain dataset is the result of a collaborative effort between the
teams of Dr. Katrin Amunts and Dr. Karl Zilles (Forschungszentrum Jülich)
and Dr. Alan Evans (Montreal Neurological Institute).
Amunts, K. et al.: "BigBrain: An Ultrahigh-Resolution 3D Human
Brain Model", Science (2013) 340 no. 6139 1472-1475, June 2013.
https://www.sciencemag.org/content/340/6139/1472.abstract
For more information please visit the BigBrain Project website.
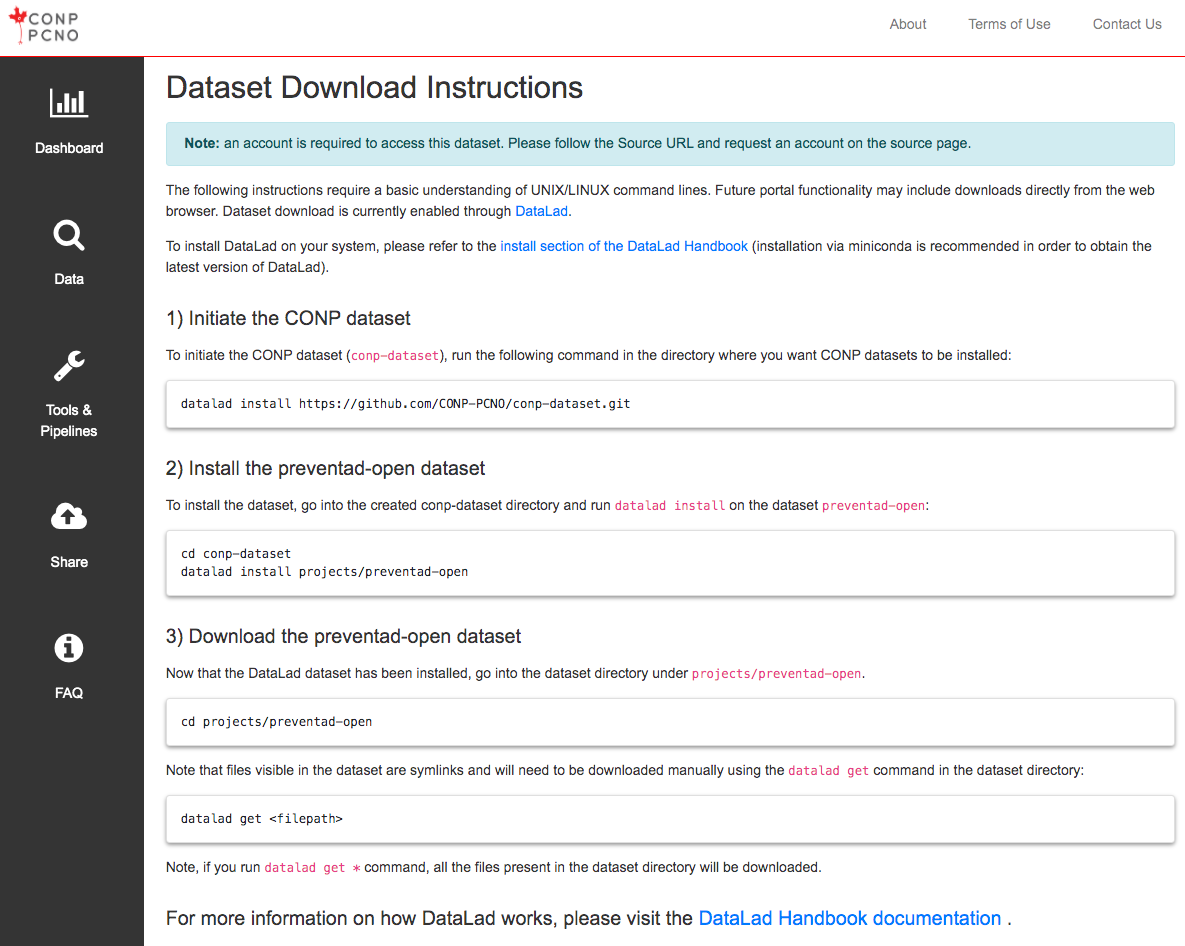
The following instructions require a basic understanding of UNIX/LINUX command lines. A subset of open datasets on the Portal are also available through a browser-based download button. The instructions below regard dataset download with the use of DataLad. To install DataLad on your system, please refer to the install section of the DataLad Handbook .
Note: For maximum compatibility with conp-dataset, the CONP recommends versions 3.12+ of Python, 10.20241202+ of git-annex, and 1.1.4+ of datalad.
1) Initiate the CONP dataset
Run the following command in the directory where you want the CONP dataset (conp-dataset) to be installed:
2) Install the BigBrain_Surface_Parcellations dataset
To install the BigBrain_Surface_Parcellations dataset, run the following commands to move into the "projects" subdirectory under the "conp-dataset" directory (created in the previous step) and run datalad install:
3) Download data from the BigBrain_Surface_Parcellations dataset
Now that the dataset has been installed, go into the BigBrain_Surface_Parcellations dataset directory.
The files visible after installing the dataset but before downloading (in the next step) are symbolic links and need to be downloaded manually using the datalad get command:
If you run datalad get * command, all the files available in the dataset directory will be downloaded.
 Experiments - Beta
Experiments - Beta
