CONP Portal | Dataset
Calgary Campinas Brain MRI Dataset
| Acknowledges: | Canada Open Neuroscience Platform, Hotchkiss Brain Institute |
|---|
Description:
Dataset README information
Calgary-Campinas Public Brain MR Dataset
Thank you for your interest in the Calgary-Campinas (CC) Public Brain Magnetic Resonance (MR) imaging dataset.
The dataset is currently composed of 3D, T1-weighted reconstructed brain MR images and segmentation masks for certain structures. It also has brain MR raw data (i.e., k-space).
Dataset Folder Structure
CC359/
└── Reconstructed
│ ├── hippocampus_staple.zip -> Silver-standard hippocampus maks generated using STAPLE
│ ├── hippocampus_subfields_freesurfer.zip -> Hippocampus subfileds masks obtained with FreeSurfer 6.0
│ ├── Manual-BEaST-mask-correction.zip -> Manual brain segmentation obtained by fixing BEaST masks (12 subjects)
│ ├── Manual.zip -> Manual brain segmentation (12 subjects)
│ ├── Original.zip -> Original images
│ ├── Silver-standard-machine-learning.zip -> Silver-standard brain maks generated using a machine learning method
| └── Silver-standard-STAPLE.zip -> Silver-standard brain maks generated using STAPLE
│
└── Raw-data
├── Multi-channel
│ ├── 12-channel
│ │ ├── test_12_channel.zip -> Undersampled 12-channel test set for R = 5 and R = 10
│ │ └── train_val_12_channel.zip -> Fully sampled 12-channel train and validation data
│ └── 32-channel
│ └── test_32_channel.zip -> Undersampled 32-channel test set for R = 5 and R = 10
└── Single-channel (Simulated single-channel raw MR data - DISCONTINUED)
├── Train_part1.zip
├── Train_part2.zip
├── Train_part3.zip
└── Val.zipReconstructed Data: Multi-vendor, multi-field-strength T1-weighted brain MR Images
The reconstructed dataset is intended for developing data-driven automatic segmentation methods and age prediction methods. At the moment, this dataset is composed of healthy brain images along with their brain masks "silver-standards" generated both using the STAPLE algorithm [1] and using supervised classification. We also provide hippocampus "silver-standards" generated using STAPLE and the hippocampus subfields masks obtained using FreeSUrfer 6.0. We are prioritizing "silver-standards" generated with STAPLE as opposed to machine learning techniques to avoid having to (re-)train algorithms for every new structure that we include in the dataset, and also because results for STAPLE and machine learning seem to be within inter-rater variability.
- The original images naming convention is the following: .nii.gz
- The skull-stripping-masks silver standards mask were generated from HWA, ROBEX, BET, OPTIBET, ANTs, BSE, MBWSS, BEaST
- The hippocampus silver standard mask were generated from FreeSurfer, FSL, Hipodeep automatic segmentation methods. There are 357 masks. CC0303 and CC0309 failed.
- The hippocampus subfields were computed with FreeSurfer 6.0. The naming convention is _h_subfields.nii.gz (: l for left hemisphere or r for right hemisphere)
Raw MR Data
Single-channel Coil Data (~8 GB uncompressed - DISCONTINUED)
We are providing 35 fully-sampled (+ 10 for testing) T1-weighted MR datasets acquired on a clinical MR scanner (Discovery MR750; General Electric (GE) Healthcare, Waukesha, WI) . Data were acquired with a 12-channel imaging coil. The multi-coil k-space data was reconstructed using vendor supplied tools (Orchestra Toolbox; GE Healthcare). Coil sensitivity maps were normalized to produce a single complex-valued image set that could be back-transformed to regenerate complex k-space samples. You can see this data as a 3D acquisition in which the inverse Fourier Transform was applied on the readout direction, which essentially allows you to treat this problem as a 2D problem while at the same time undersampling on two directions (slice encoding and phase encoding). The matrix size is 256 x 256.
We are providing the train and validation sets. The data are split as follows:
Train: 25 subjects - 4,524 slices
Validation: 10 subjects - 1,700 slices
Test: 10 subjects - 1,700 slices (not provided, it will be used to test the model you submit)
This synthetic single-coil data is meant as proof of concept for assessing the ability of using Deep Learning for MR reconstruction. If you already have experience with that, we suggest that you go straight to the multi-channel raw data.
Multi-channel Coil Data (~219 GB uncompressed)
We are providing 167 three-dimensional (3D) , T1-weighted, gradient-recalled echo, 1 mm isotropic sagittal acquisitions collected on a clinical MR scanner (Discovery MR750; General Electric (GE) Healthcare, Waukesha, WI). The scans correspond to presumed healthy subjects (age: 44.5 years +/- 15.5 years [mean +/- standard deviation]; range: 20 years to 80 years). Datasets were acquired using either a 12-channel (117 scans) or a 32-channel coil (50 scans). Acquisition parameters were TR/TE/TI = 6.3 ms/ 2.6 ms/ 650 ms (93 scans) and TR/TE/TI = 7.4 ms/ 3.1 ms/ 400 ms (74 scans), with 170 to 180 contiguous 1.0-mm slices and a field of view of 256 mm x 218 mm. The acquisition matrix size for each channel was Nx x Ny x Nz = 256 x 218 x [170,180]. In the slice-encoded direction (kz), data were partially collected up to 85% of its matrix size and then zero filled to Nz= [170,180]. The scanner automatically applied the inverse Fourier Transform, using the fast Fourier transform (FFT) algorithms, to the kx-ky-kz k-space data in the frequency-encoded direction, so a hybrid x-ky-kz dataset was saved. This reduces the problem from 3D to two-dimensions, while still allowing to undersample k-space in the phase encoding and slice encoding directions. The partial Fourier reference data were reconstructed by taking the channel-wise iFFT of the collected k-spaces for each slice of the 3D volume and combining the outputs through the conventional sum of squares algorithm. The dataset train/validation/test split is summarized in the table below .Relevant information
- Healthy subjects (age: 44.5 years ± 15.5 years; range: 20 years to 80 years).
- Acquisition parameters are either: TR/TE/TI = 6.3 ms/2.6 ms/650 ms and TR/TE/TI = 7.4 ms/3.1 ms/400 ms
- Average scan duration ~341 seconds
- Only the undersampled k-spaces for R=5 and R=10 are provided for the test set
Dataset summary:
- 12-channel
- Train: 47 dataset
- Validation: 20 datasets
- Test: 50 datasets
- 32-channel
- Test: 50 datasets
License
Our datasets are distributed under a Creative Commons Attribution-NoDerivatives 4.0 International Public License.
Contact information
roberto.medeirosdeso@ucalgary.ca (dataset manager)
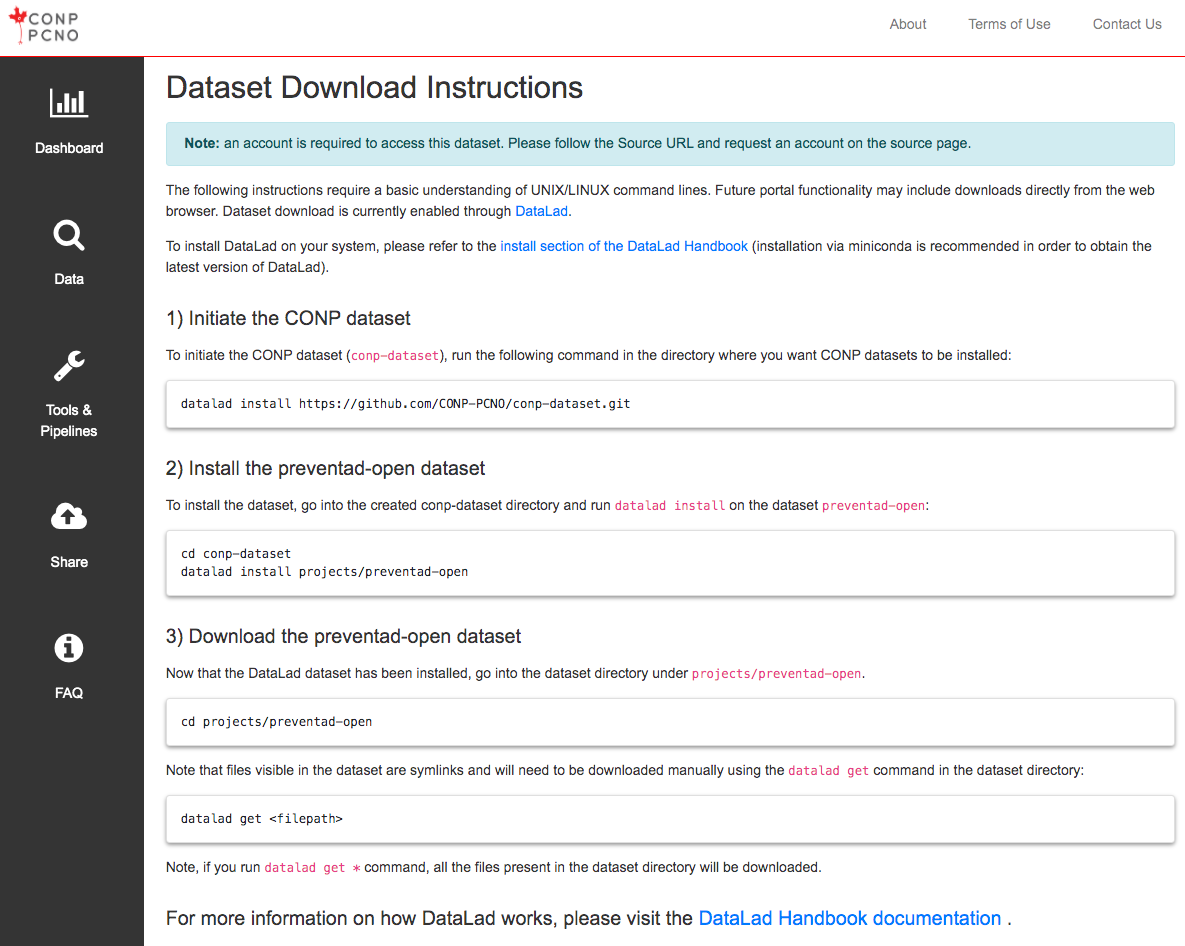
The following instructions require a basic understanding of UNIX/LINUX command lines. A subset of open datasets on the Portal are also available through a browser-based download button. The instructions below regard dataset download with the use of DataLad. To install DataLad on your system, please refer to the install section of the DataLad Handbook .
Note: For maximum compatibility with conp-dataset, the CONP recommends versions 3.12+ of Python, 10.20241202+ of git-annex, and 1.1.4+ of datalad.
1) Initiate the CONP dataset
Run the following command in the directory where you want the CONP dataset (conp-dataset) to be installed:
2) Install the calgary-campinas dataset
To install the calgary-campinas dataset, run the following commands to move into the "projects" subdirectory under the "conp-dataset" directory (created in the previous step) and run datalad install:
3) Download data from the calgary-campinas dataset
Now that the dataset has been installed, go into the calgary-campinas dataset directory.
The files visible after installing the dataset but before downloading (in the next step) are symbolic links and need to be downloaded manually using the datalad get command:
If you run datalad get * command, all the files available in the dataset directory will be downloaded.
 Experiments - Beta
Experiments - Beta
