(Last Updated on January 20, 2021)
This Privacy Policy governs the collection, use, and disclosure of information generated during your visit
to the Canadian Open Neuroscience Platform (CONP) Portal. The Portal is hosted by the McGill Centre for
Integrative Neuroscience (MCIN), a unit within McGill University.
This Policy is separate from the Portal’s Terms of Use, Data Contributor Agreement, or other CONP-related
materials.
Definitions
For the purposes of this Policy:
“aggregate-level data” refers to data that has been grouped together to provide information at a broader
level than the initial record-level data. This data does not allow for the identification of individual
records. (Adopted from definitions by
Statistics Canada
, the
Canadian Institute for Health Information
, and the
Organisation for Economic Co-operation and Development
.)
“asset” refers to either a dataset, tool, or pipeline available through the CONP Portal.
“CONP” refers to the Canadian Open Neuroscience Platform.
“cookie” is a small electronic file temporarily stored on your computer that contains information about your
visit to a website.
“external website” refers to any website other than that accessible at
https://portal.conp.ca/.
“MCIN” refers to the McGill Centre for Integrative Neuroscience, which runs the CONP Portal.
“personal information” refers to individually identifying information for which there is a reasonable
likelihood of identification.
“Portal” refers to the web interface for the CONP Portal, which is accessible at
https://portal.conp.ca/
.
“public IP address” means a unique address that publicly identifies your connection on the Internet.
“Terms of Use” refers to the user agreement that governs your use of the resources available on the CONP
portal. It is distinct from this Policy.
Consent Statement
By visiting the Portal, you agree to the terms of this Policy. By your use of the Portal, you consent to the
collection, use, and disclosure of your personal information in accordance with the terms contained in this
Policy, which may be amended from time to time. You understand that it is your responsibility to verify any
updates to this Policy.
Collection of Personal Information
Information about Your Visits
CONP uses the software
Matomo
to gather information about your visits. Thanks to Matomo, this information does not leave CONP servers,
allowing CONP to maintain control of this data. When you visit and use the Portal, our server automatically
collects certain technical information pertaining to your visit and use of the Portal. These include:
- your public IP address and its approximate geolocation
- the domain name from which you visited the Portal
- user-specific information on which pages are visited
- aggregate information on pages visited
- the date and time of visit
- asset accessed on the Portal
We use this information to improve the Portal and for reporting purposes. We do not combine this information
with that from other sources to identify individual users. This technical information is retained for up to
12 months before being aggregated.
User-Submitted Survey Information
CONP uses Google Forms to survey Portal users about their experiences using the Portal. This information is
not automatically collected and any user information submitted via the Google Form is entirely of the user’s
choosing.
Where a Portal user chooses to submit feedback via Google Forms, the information is subject to Google’s own
Privacy Policy. See also the section “Links to External Websites” below.
Information Sharing
CONP publishes aggregate-level data about the use of the Portal both on the Portal website and in reports
related to CONP’s activities.
CONP does not share any personal information with other entities or organizations, except when legally
required to do so, at the request of governmental authorities conducting an investigation, to verify or
enforce compliance with the policies governing the Portal and applicable laws, or to protect against misuse
or unauthorized use of the Portal.
Except as described above, we will not share any information with any party for any reason.
Cookies
We use cookies to:
- Identify your visit to the Portal
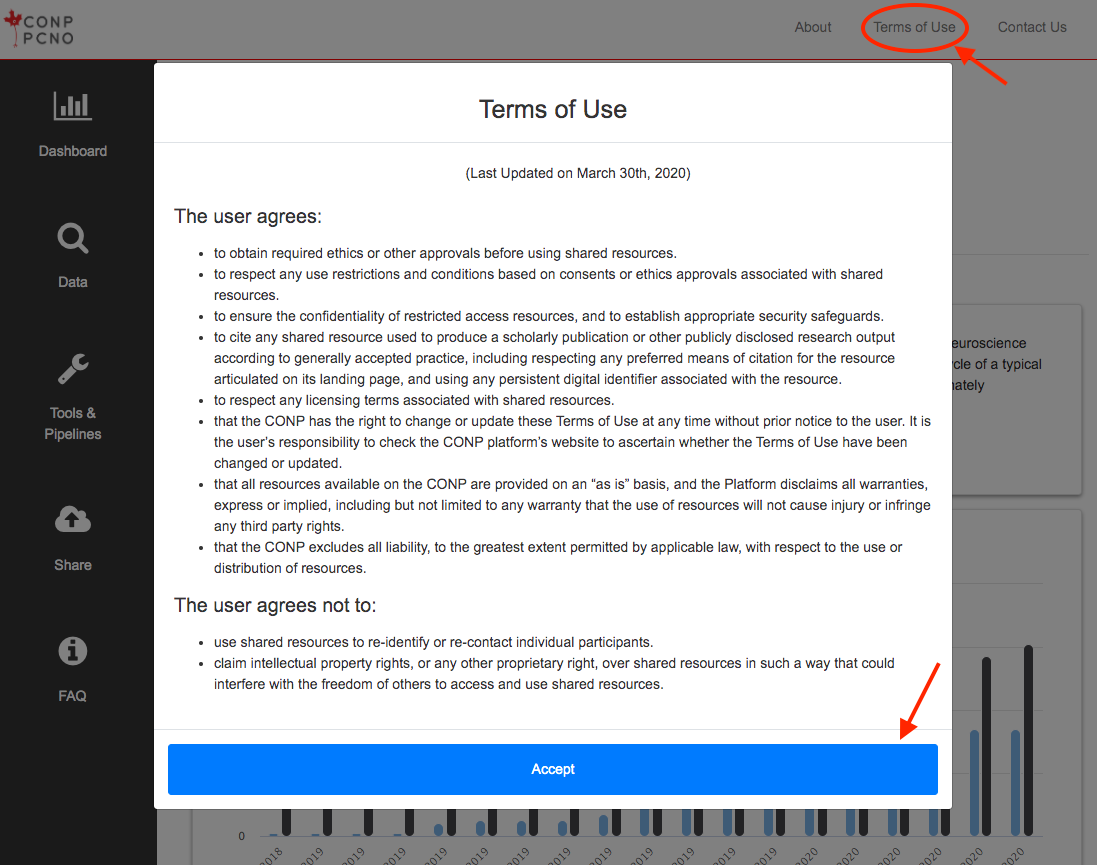
- Record your acceptance of the Terms of Use
As acceptance of the Terms of Use is a prerequisite to accessing the site, it is not possible to block this
cookie and maintain site access.
Disclosure of Personal Information
In so far as the information collected is considered personal information, and within the custody of MCIN,
it will not be shared with individuals and units outside of MCIN. We will not disclose personally
identifiable information about your use of the site except under the following circumstances:
- With your prior explicit consent
-
When we have given you clear notice that we will disclose information that you voluntarily provide
-
With appropriate external parties, such as law enforcement agencies, in order to investigate and respond
to suspected violations of law or university policy. Any such disclosures shall comply with all applicable
laws and institutional policies. Only that information which is necessary for compliance purposes will be
disclosed.
Security
Due to the rapidly evolving nature of information technologies, no transmission of information over the
Internet can be guaranteed to be completely secure. While CONP is committed to protecting user privacy, CONP
cannot guarantee the security of any information users transmit to university sites, and users do so at
their own risk.
MCIN staff, who run the CONP Portal, are subject to McGill University’s
Policy on the Responsible Use of McGill Information Technology Resources
. Pursuant to this policy, we have appropriate organizational and technical security measures in place in
our physical facilities to protect against the loss, misuse, or alteration of information that we have
collected from you at our site. We furthermore use reasonable safeguards consistent with prevailing industry
standards and proportionate to the sensitivity of the data being stored to maintain the security of that
information on our systems.
Links to External Websites
The Portal contains links to external websites. CONP is not responsible for the availability, content, or
privacy practices of external websites. These external websites have their own policies as it relates to the
collection, use, and disclosure of personal information. We invite you to consult the privacy policies of
these websites to know more.
Privacy Notice Changes
From time to time, we may use visitor information for new, unanticipated uses not previously disclosed in
this Privacy Policy. In such cases, this Privacy Policy will be updated accordingly. If this is a concern,
we encourage you to periodically check for changes.
Questions about this Policy
Should you have any questions or concerns about this Privacy Policy, kindly send us an email at
privacy@conp.ca
.
 Experiments - Beta
Experiments - Beta