CONP Portal | Dataset
MICA-MICs: a dataset for Microstructure-Informed Connectomics
| Acknowledges: | Canadian Institute of Health Research , Canadian Open Neuroscience Platform, Canada Research Chairs program, National Sciences and Engineering Research Council of Canada |
|---|
Description:
Dataset README information
MICA-MICs: a dataset for Microstructure-Informed Connectomics
The human brain is a highly interconnected network which can be described at multiple spatial and temporal scales. Neuroimaging, in particular magnetic resonance imaging (MRI), has provided a window into brain structure and function, offering versatile contrasts to assess its multiscale organization. Here, we leverage the rich descriptions of brain structure and function offered by MRI to further our understanding of human brain organization across modalities and spatial scales. MICA-MICs is a dataset for microstructure-informed connectomics providing raw and fully processed multimodal neuroimaging data acquired in 50 healthy control participants at an MRI field strength of 3T. Modalities include high-resolution anatomical (T1-weighted), microstructurally-sensitive (quantitative T1), diffusion-weighted, and resting-state functional imaging. We additionally provide users with ready-to-use connectomes built across multiple parcellation schemes based on histology (e.g. Von Economo), sulco-gyral landmarks (e.g. Desikan-Killiany), and function (e.g. Schaefer atlases), for a total of 18 different parcellations of varying spatial scale. All connectome matrices were generated using micapipe (https://micapipe.readthedocs.io/), a robust pre-processing pipeline for multimodal neuroimaging data. By granting access to data modalities at different pre-processing stages, this dataset will streamline the development of novel approaches to study complex interactions between regional and networks-level properties of the human brain.
Please cite the following reference if you use this dataset:
Royer, J., Rodriguez-Cruces, R., Tavakol, S., Lariviere, S., Herholz, P., Li, Q., Vos de Wael, R., Paquola, C., Benkarim, O., Park, B., Lowe, A.J., Margulies, D.S., Smallwood, J., Bernasconi, A., Bernasconi, N., Frauscher, B., Bernhardt, B.C., 2021. An open MRI dataset for multiscale neuroscience. bioRxiv 2021.08.04.454795. https://doi.org/10.1101/2021.08.04.454795
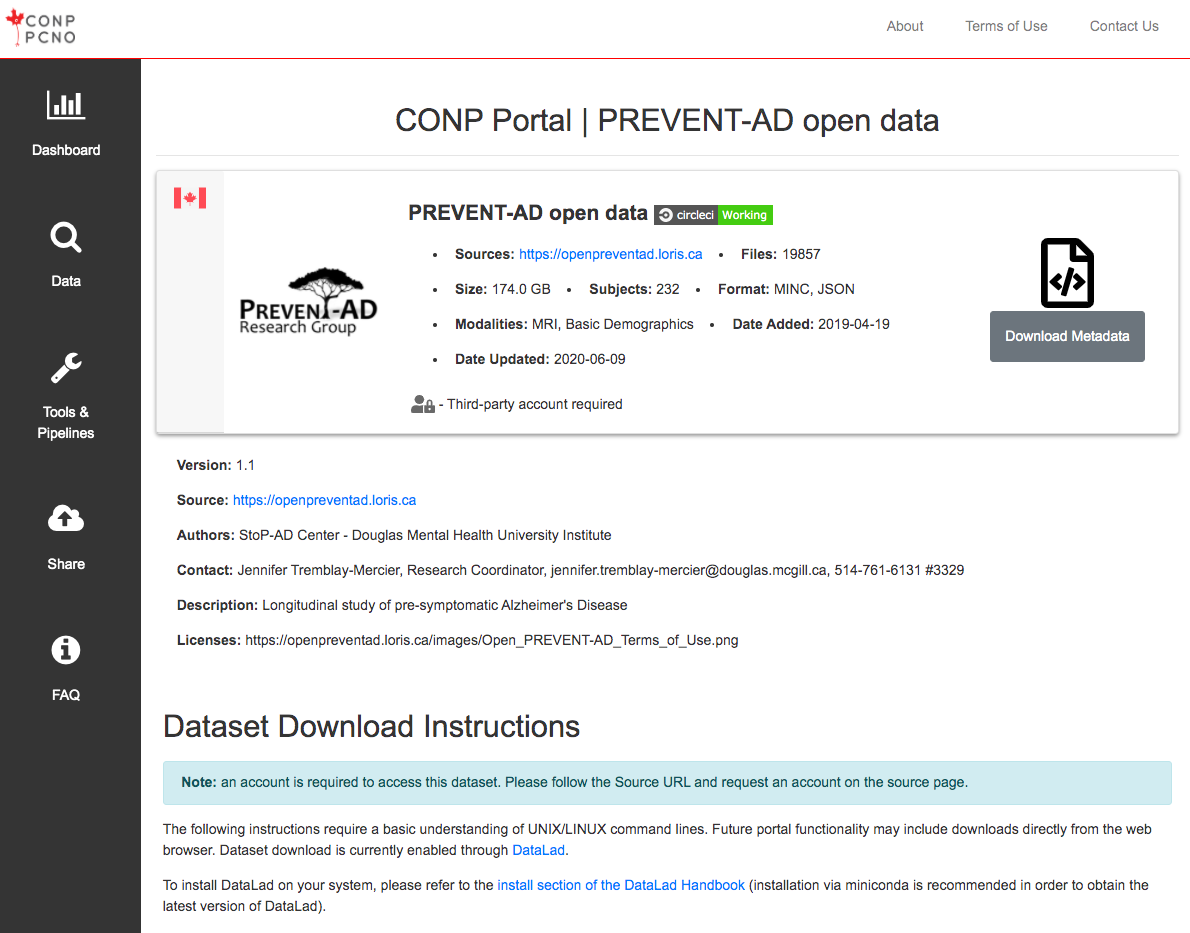
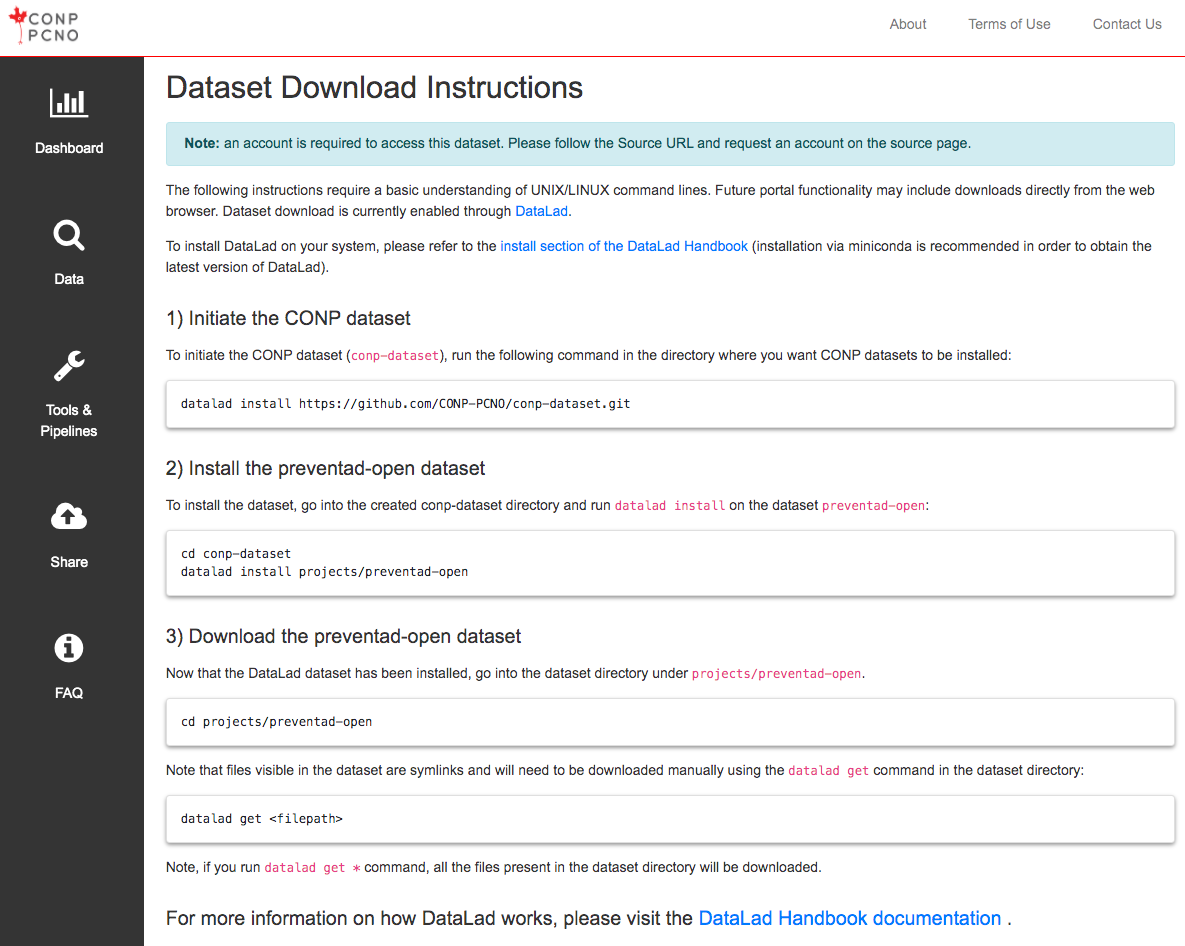
The following instructions require a basic understanding of UNIX/LINUX command lines. Future portal functionality may include downloads directly from the web browser. Dataset download is currently enabled through DataLad.
Note: The conp-dataset requires version >=0.12.5 of DataLad and version >=8.20200309 of git-annex.
To install DataLad on your system, please refer to the install section of the DataLad Handbook (installation via miniconda is recommended in order to obtain the latest version of DataLad).
1) Initiate the CONP dataset
To initiate the CONP dataset (conp-dataset), run the following command in the directory where you want
CONP datasets to be installed:
datalad install https://github.com/CONP-PCNO/conp-dataset.git
2) Install the mica-mics dataset
To install the dataset, go into the created conp-dataset directory and run
datalad install on the dataset mica-mics:
cd conp-dataset
datalad install projects/mica-mics
3) Download the mica-mics dataset
Now that the DataLad dataset has been installed, go into the dataset directory under
projects/mica-mics.
cd projects/mica-mics
Note that files visible in the dataset are symlinks and will need to be downloaded manually using the
datalad get
command in the dataset directory:
datalad get <filepath>
Note, if you run datalad get * command, all the files present in the dataset directory will be
downloaded.